Genetic information in all life cells is kept within the primary sequences of DNA and RNA molecules. Both of them are heteropolymers consisting of four different nucleotide types. The question « why nature uses exactly four letters to encode genetic information » was discussed since the role of DNA and RNA in storing and transmission of genetic information has been understood. Typically, the attempts to answer it are based on the chemistry of interacting nucleotides, or deal with the conjectures lying in the general information theory. We recently presented a new observation concerning the statistics of RNA secondary structures. This observation may be a new contribution to the problem of « why only four? ».
The RNA molecules are known to form secondary structures (i.e. intra-molecular structures stabilized by theromoreversible hydrogen bonds between non-neighboring nucleotides) which mostly take a « cactus-like » (or « cloverleaf-like ») hierarchically folded form, topologically isomorphic to a tree:
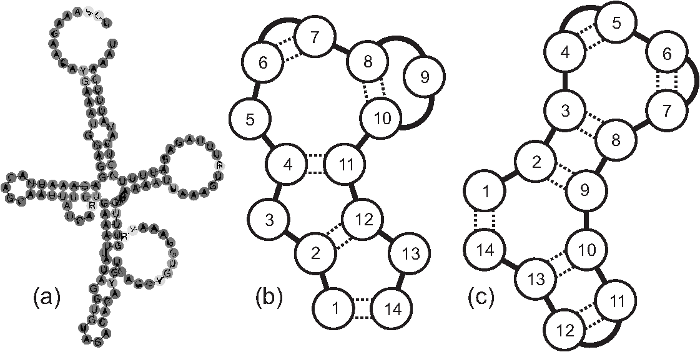
(b) and (c) Schematic cloverleaf structures of RNA-like chains with and without gaps respectively.
Using the methods of statistical physics we demonstrate the existence of a specific morphological transition which occurs in a toy model of random RNA-like chains depending on the number of nucleotide types (letters) c used to construct the chain. This morphological transition occurs at ccr=4 (i.e., at four nucleotide types), making this particular number of nucleotide types special. The transition has a very transparent physical meaning: for ccr in the thermodynamic limit (i.e. for very long chains) there exists a gapless « perfect » secondary structure, that is a structure in which the fraction of nucleotides connected to the complimentary ones via hydrogen bonds equals one, while for c>ccr even in the best possible secondary structure there always exists a finite fraction of gaps (i.e., nucleotides which have nobody to connect with).
Such a criticality is specific only for RNA chains and is due to the additional freedom existing in the formation of the complex cactus-like secondary structures. For DNA chains the fraction of matched nucleotides in the optimal alignment of two random sequences is less than 1 for any alphabet with c>1. Thus, the exclusivity of RNA 4–letter alphabets is consistent with the modern opinion that the origin of life could be connected with the template–directed replication of random RNA molecules (the so-called « RNA world » hypothesis).
See O.V. Valba, M.V. Tamm and S.K. Nechaev, On exclusivity of alphabets with four nucleotide type, arxiv.org/abs/1109.5410
